Research
Infertility afflicts 10% of couples in the United States, of which half are attributable to male factors. Of these, one third are thought to be due to defects in sperm production leading to morphologically abnormal or poorly motile sperm. It has long been appreciated that the transcriptome of spermatogenic cells is exquisitely regulated during the sequential progression from spermatogonial stem cell to mature spermatozoa, but few studies to date have systematically investigated this regulation. Indeed, the male germ cell appears to establish particularly complex patterns of transcriptional and post-transcriptional regulation that extends to many levels of the mRNA life cycle, including testis-specific transcription; alternative splicing, poly-adenylation and use of 3′ untranslated regions (3′UTR); and the deposition of methylation marks on RNA (known as epitranscriptomic regulation). Importantly, all these levels of RNA metabolism share one alarming feature relating to male fertility: human variants in the genes regulating each of these steps are associated with aberrant sperm morphology and infertility in mice and in men. These observations demonstrate that stringent regulation of gene expression throughout spermatogenesis is critical for the production of healthy spermatozoa.
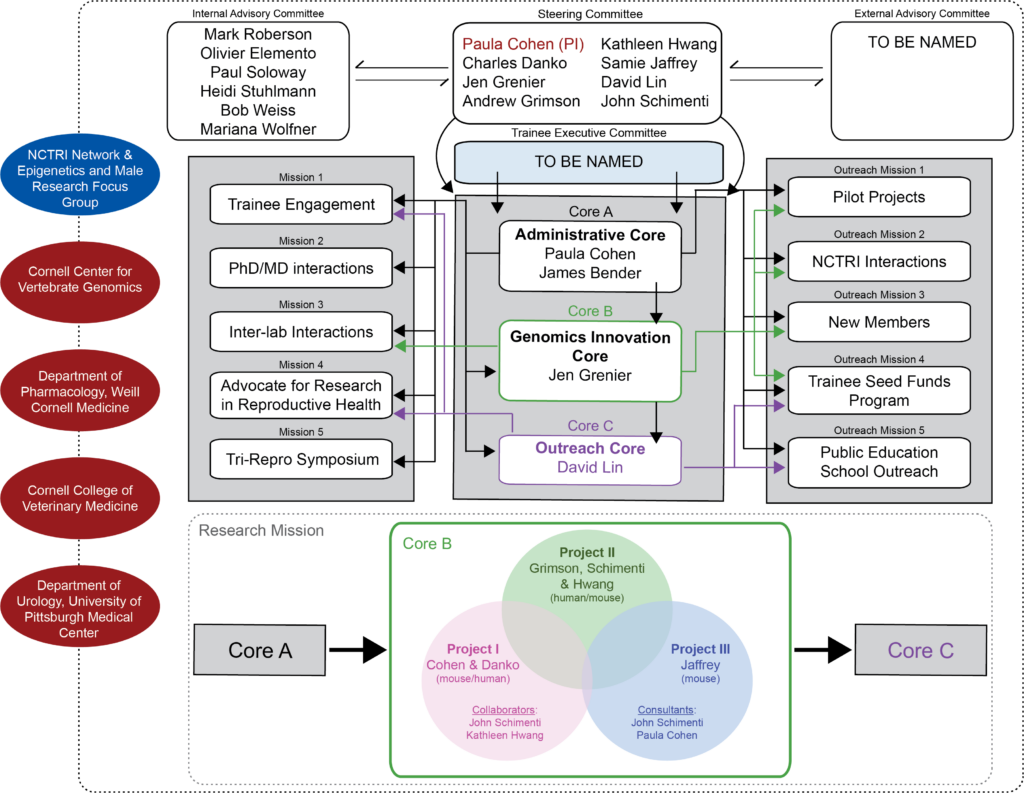
The current P50 is aimed at defining how transcriptional, post-transcriptional, and epitranscriptomic control of spermatogenesis is achieved and coordinated to produce a defined pattern of gene expression, at the same time ensuring appropriate chromatin compaction events during spermiogenesis. Interwoven throughout this proposal is a strong translational focus to ask how we can use the basic biology to define new benchmarks and criteria by which to judge the fertilization potential of sperm from men with a high rate of sperm morphological abnormalities. Our studies build upon the unique strengths and expertise in spermatogenesis and gene regulation within the CoRe, along with an exciting new collaboration with the University of Pittsburgh. Three projects are proposed, as described below along with three accompanying core facilities that support all component labs of the P50.
Primarily, all component projects are supported by a strong ADMINISTRATIVE CORE (Leader: Paula Cohen) that will facilitate close interactions through regular meetings, trainee events, and the “Tri-Repro” Annual Symposium that attracts participants nationwide, and which has recently joined forces with similar P50 centers at The Universities of Pennsylvania and Pittsburgh. This core will provide trainees with opportunities to improve their grantsmanship through a Seed Grant competition, whilst larger Pilot Projects will allow for the recruitment of new CoRe faculty with interests in reproduction. Our state-of-the-art GENOMIC PROFILING CORE (Leader: Jen Grenier) will serve as an Innovation Hub for exploring all aspects of gene regulation in reproduction, specializing in a range of next generation sequencing technologies including RNA-seq, ATAC-seq, and PRO/ChRO-seq, as well as cutting-edge combinatorial indexing approaches for ultra-high throughput and single-cell profiling applications. Finally, our OUTREACH CORE (Leader: David Lin) will target rural school districts in upstate New York, a traditionally underserved community, by engaging school children in scientific activities organized by faculty and trainees. As well as providing mentorship and insight into scientific research as a career, we will create innovative new hands-on activities focused on principles of reproductive biology, illustrating principles of reproduction at the chromosomal, cellular, organismal, and evolutionary levels.
All these components of our center will benefit from the strong foundations and collaborative integration we have established over the past 13 years, and by robust and unequivocal institutional support and resources.
Cohen and Danko Labs
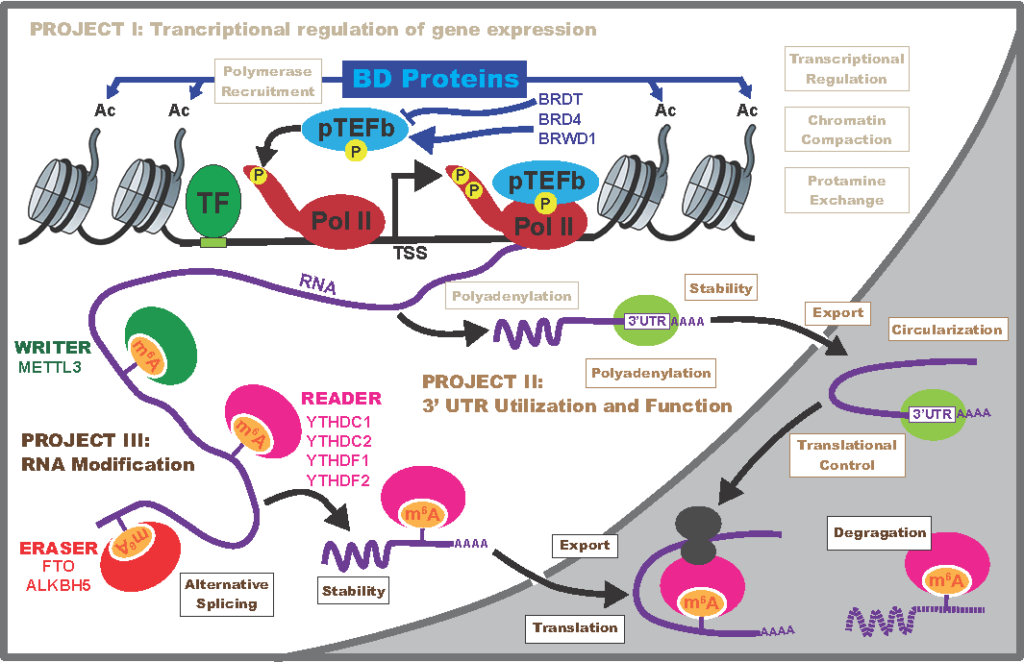
PROJECT I (Leaders: Paula Cohen and Charles Danko) explores the clinical significance of transcriptional control of gene expression during spermatogenesis focusing on the mechanism of transcriptional shutdown in the transition from meiosis to spermiogenesis. Specifically, this project will investigate the integrated function of bromodomain proteins in orchestrating the unique and stage-specific transcriptome, their role in 3’UTR utilization and polyadenylation site selection, and the impact of bromodomain disruption for human spermatogenesis.
Grimson and Schimenti Labs
PROJECT II (Leaders: Andrew Grimson, John Schimenti, and Kathleen Hwang) will explore the importance of post-transcriptional control of gene expression for production of healthy spermatozoa, and specifically focuses on the role of the 3′ untranslated region as the major source of post-transcriptional regulatory information. Using morphologically characterized sperm populations from fertile and infertile men, this study seeks to identify pathogenic 3’UTR variants that affect expression or levels of key spermiogenesis mRNAs in ways that impact fertility and sperm function. High throughput assays and mouse modeling will be employed to these ends.
PROJECT III (Leader: Samie Jaffrey) is based on the finding that epitranscriptomic modifications of mRNA, particularly those involving N6-methyladenosine (m6A) and N6, 2’-O-dimethyladenosine (m6Am), play a major role in regulating 3′UTR sequence utilization and protein translation during spermatogenesis. This project will test the hypothesis such modifications might explain some of the unusual properties of mRNA stability and translational delay during spermatogenesis. Studies in this project will focus on the role of m6A modifications on selection of germ cell-specific 3′UTR sequences, the role of epitranscriptomic regulation of long non-coding RNAs (lncRNA) in regulating sperm development, and the role of m6Am modifications in controlling spermatogenesis.
External Advisory Committee
The External Advisory Committee (EAC) consists of stellar researchers from around the country with expertise in areas of research that are highly related to the goals of our P50. The EAC advises the P50 participants on research directions and assists in evaluating new Pilot projects, while at the same time helping to better appreciate how an understanding of gene regulation in the germ cell can assist in our knowledge and treatment of human infertility as it pertains to sperm quality. EAC members participate in our Annual Tri-Repro Symposium where they will be apprised of all Center activities over the preceding year in the form of poster and oral presentations from P50 participants, as well as during a joint session of the EAC, IAC and P50 faculty.

Jackson Laboratory, Bar Harbor, ME

Memorial Sloan Kettering Cancer Center, New York, NY

Cold Spring Harbor Laboratory, Laurel Hollow, NY

University of Pennsylvania, Philadelphia, PA

Johns Hopkins Medicine, Baltimore, MD
Internal Advisory Committee
The Internal Advisory Committee (IAC) for our P50 grant oversees all activities of the Center Grant components, meets regularly with the project/core leaders of the P50, and attends biannual meetings to evaluate the progress of the research. They work closely with the Principal Investigator to ensure that the goals of the grant are being met, and they serve as a consulting group for any conflicts or difficulties being experienced by P50 participants.

Weill Cornell Medicine
Dr. Elemento is a Professor of Physiology at Weill Cornell Medicine and the Director of the Caryl and Israel Englander Institute for Precision Medicine. Dr. Elemento is an outstanding computational biologist with expertise in big data, single cell technologies, and development of analysis algorithms.

College of Veterinary Medicine
Dr. Soloway is Chair of the Department of Biomedical Sciences and a leading authority on epigenetic imprinting. His research focuses on mechanisms regulating epigenetic states, and developing single molecule approaches for epigenomic analysis. As chair of the host department for CoRe, Dr. Soloway has maintained a keen interest in the activities of our center.

Weill Cornell Medicine
Dr. Stuhlmann is a Professor of Cell and Developmental Biology and a Professor of Cell and Developmental Biology in Pediatrics. She is also an Adjunct Professor in the Department of Biomedical Sciences at the Cornell University Veterinary College. Since 2021 she serves as the Interim Chair of the Departments of Biochemistry and Cell and Developmental Biology. Her lab focuses on the molecular and genetic pathways that regulate vascular endothelial lineage specification, and on regulatory and epigenetic mechanism in placental development and disease.

College of Veterinary Medicine
Dr. Weiss is a Professor of Genetics in the Department of Biomedical Sciences, and the Associate Dean for Research at the College of Veterinary Medicine. A leader in the field of DNA damage repair, Dr. Weiss studies the physiological roles for the DNA damage checkpoint protein Hus1, a component of the RAD9-RAD1-HUS1 (911) complex, in mammalian germ cells. Dr. Weiss has been a member of the CoRe since 2010.

College of Arts and Sciences
Dr. Roberson is a Professor of Biomedical Sciences, and the former chair of this department. He has served as chair of our CoRe Oversight Committee since 2007 and currently serves as Chair of our IAC for this P50. He is a leading expert on the on the neuroendocrine pathways that regulate reproduction, and on the molecular mechanisms that promote placentation.



